@fjaviersanchez: I now have a solution that I hope will work for you. It needs some more work, but I thought I’d put it out there now so I can get comments. This solution makes it fairly trivial to ingest arbitrary files into LSST data repositories so you can treat them like any other image in the stack. In order to use it, you’ll need the tickets/DM-6924 branch of both obs_file and pipe_tasks.
To run processFile is a three step process. The first two are one time only if your input data are static. The following assumes you have a relatively recent master build and the ticket branches setup.
- Create a repository to feed the ingest. This involves making a directory containing a file called
_mapper. I made a directory called test_out. The content of the file is the single line lsst.obs.file.FileMapper.
- Ingest the files. This puts the files in the location expected by the rest of the machinery and builds a database to lookup the available images.
$> ingestFiles.py test_out test_imgs/test.fits.gz
The first argument is a valid repository and the rest of the arguments will be treated as images to ingest. If no output is specified the images will be ingested into the repository specified (i.e. test_out).
- Run processCcd
$> processCcd.py test_out/ --id filename='test.fits.gz' --config isr.noise=100000 isr.addNoise=True --output test_out
Just specify the file you want to process. In the case of your data, I also had to add background noise, otherwise certain algorithms, e.g. cosmic ray detection, do not behave well. The output argument is required.
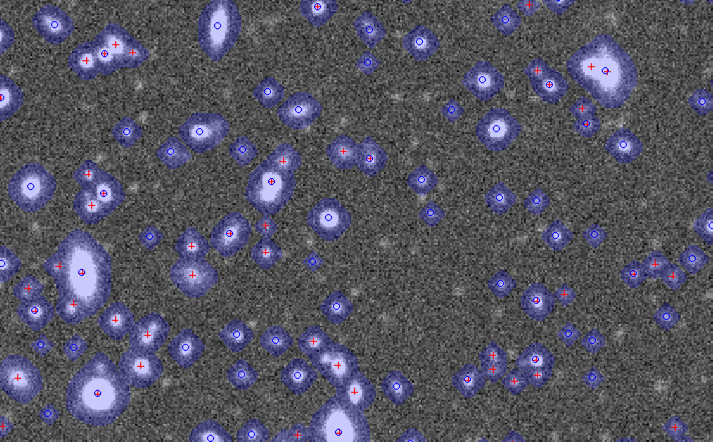
Below is an example produced by the displaySources.py utility. If you have display_ds9 setup, you can specify a repository and a file and it will plot the parent sources as blue circles and the children as red plusses. Looking at this image, there are some things to tune. For example, I see some faint sources I would have expected to be detected, but maybe that is an artifact of my arbitrary choice of background noise. FYI, the blue patches are the pixels flagged as detected.